How to prepare low yield (< 2 nM) libraries for sequencing on a MiSeq
The MiSeq Denature and Dilute Libraries Guide has directions for denaturing 2 nM and 4 nM libraries with the Standard Normalization Method. The denaturation steps make sure that the concentration of NaOH is no more than 1 mM in the final solution diluted with HT1. Higher NaOH concentrations can inhibit library hybridization to the flow cell, leading to decreased cluster density.
Libraries that are less than 2 nM but at least 0.5 nM can be prepared for MiSeq sequencing. This is done by following the Standard Normalization Method in the NextSeq 500 and 550 Sequencing Systems Denature and Dilute Libraries Guide until libraries are diluted to 20 pM. In this protocol, excess NaOH is offset by adding 200 mM Tris-HCl to make sure that NaOH is fully hydrolyzed in the final solution.
To denature < 2 nM libraries, follow the steps for starting library concentration of 1 nM or 0.5 nM in the NextSeq 500 and 550 Sequencing Systems Denature and Dilute Libraries Guide, see Figure 1. When libraries are at 20 pM, proceed to the Dilute Denatured 20 pM Library step in the MiSeq System Denature and Dilute Libraries Guide, see Figure 2.
Refer to the specific Denature and Dilute Libraries Guides for specific instruments for detailed instructions.
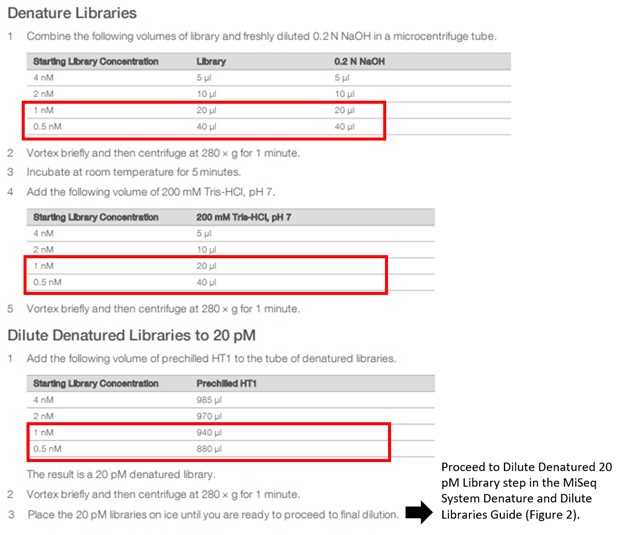
Figure 1. Denaturation and Dilution steps for Standard Normalization Method in the NextSeq 500 and 550 Sequencing Systems Denature and Dilute Libraries Guide.
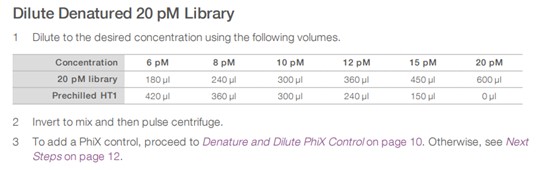
Figure 2. Dilute Denatured 20 pM Library step for the Standard Normalization Method in the MiSeq System Denature and Dilute Libraries Guide.
For any feedback or questions regarding this article (Illumina Knowledge Article #6058), contact Illumina Technical Support [email protected].
Last updated
Was this helpful?
